About NemRac
-Why we developed NemaRac?
Nematodes are ubiquitous in soil representing different trophic levels and occupying a central position in the detritus food web. Nematodes have been widely used for biomonitoring of soil quality and health. However, their application in bio-indicator is limited due to the difficulties in species identification, e.g. the high phenotypic plasticity, the vague diagnostic characters, and frequently encountered juveniles upgrade the difficult in species identification and thus a well-established experience and expertise is hence essential. Given the number of taxonomists are rapidly declining worldwide, the nematode species identification often become the bottleneck for the application of these ecological indicators.
-What NemaRac do?
In present study the CNN-based deep learning approach was used to develop an automatic nematode image-based identification and ecological indices calculation pipeline. In the test, the model can properly identify up to 60% genera, 76% of c-p values and 76% feeding types in real test dataset, and 94%-97% in simulated dataset. The pipeline was further incorporated into a user-friendly web application NemaRec. NemaRec offers high-throughput online identification while simultaneously collect images uploaded by users for future modeling training.

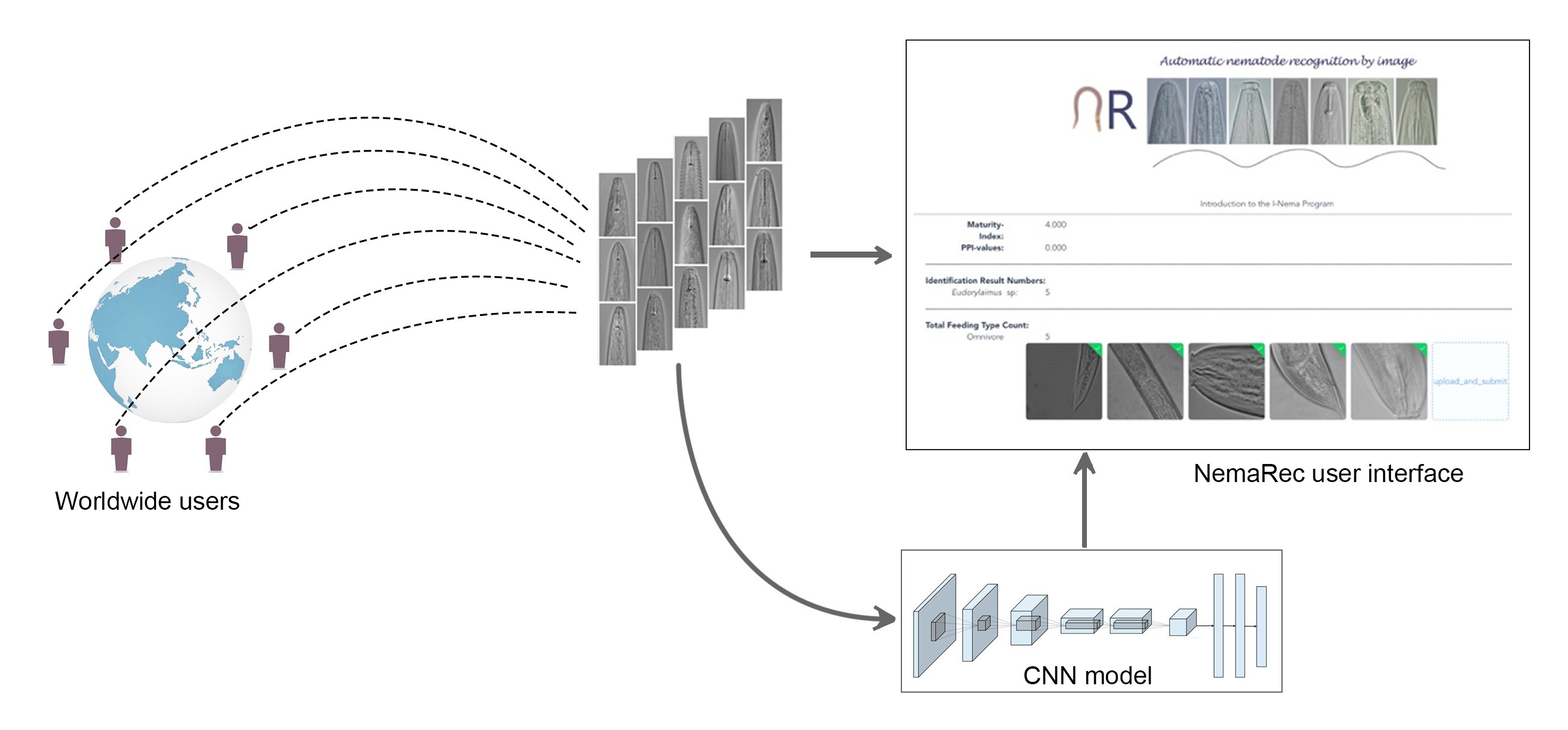
Fig. 1. The NemaRec user interface and the model re-training system by collecting user uploaded images in program back end.
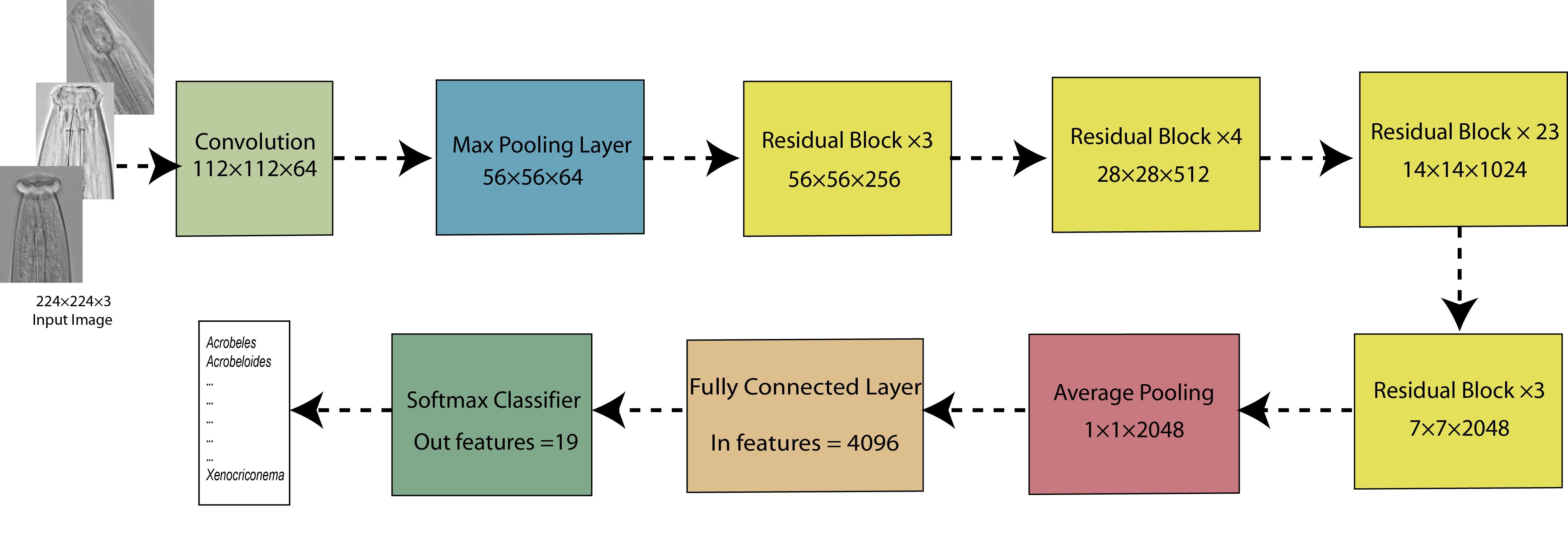
Fig. 2. The overall architecture of ResNet101 used in this study.
- How to get nematode image?
NemaRec accept images taken from microscopy, ideally at magnification of 10X16 to capture whole body and 100X16 to capture head region. Images can be save as JPEG and TIFF format, and upload in any size, but ideally less 5M.
The identification accuracy is highly dependent on the query image quality, and using high quality images can significantly improve identification success.
Here are some of our suggestions:
Try to use temporary slide for image capture, the fresh specimen can retain more the morphology characters.
Wash nematode and remove debris before making slide. Avoid debris around head and tail region.
Add sufficient water in slide, avoid crush nematode
Adjust microscopy to make sure image is taken in proper image plane.
Make sure nematodes are not crushed.
Take pictures at head region, this will give highest identification accuracy.
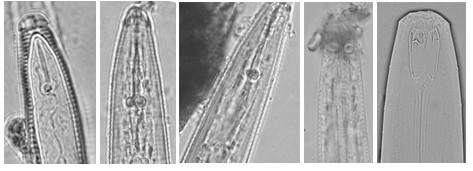
Fig 3 Example of images with low quality.
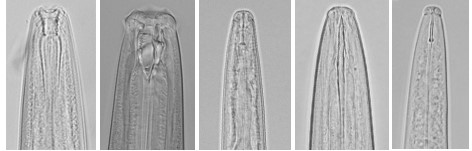
Fig 4 Example of images with high quality
-How to interpret identification results?
It is important to notice that the identification success rate is between 20-60% in current test version. This means very likely you may not have a confident result, and this method cannot replace traditional manual identification approach. The MI and PPI indices, c-p value and feeding types are calculated based on the taxa identification result. Since few taxa may share same values, MI and PPI indices, c-p value and feeding types can be accidently correct even the taxa is misidentified.
We are working on the improvement of model by adding more images for model training, and the accuracy will be more reliable in coming version.
-Copyrights
Please notice this website collects the user uploaded images for CNN model training. The copyrights are owned by their original authors and these images will not be used in any occasion other than CNN model training.
In case you don’t want your images to be collect and used for model training, please write an email to qingxue@njau.edu.cn. We will delete images from database.
-Support NemRec project
The amount of images used for model training is the main factor that affect the identification accuracy. Therefore, we encourage more researchers to join this program by providing nematode images.
You can send images through Wetransfer, Dropbox or OneDrive, to dr. Xue Qing (qingxue@njau.edu.cn). All contributors will be acknowledged in this website.
-Development team
Dr. Xue Qing
Associated professors at Nanjing Agricultural University in China
Role: Nematode images acquire and identification
Research interests: Taxonomy, phylogeny and evolution of nematodes.
Contact: qingxue@njau.edu.cn
Dr. Xuequan Lu
Assistant Professor at Deakin University in Australia
Role: Technique supports in CNN modeling training.
Research interests: Visual Data Computing, for example, geometry modeling, processing and analysis, animation/simulation, 2D data processing and analysis.
Contact: xuequan.lu@deakin.edu.au
Dr. Xiaojun Xie
Lecturer at Nanjing Agricultural University in China
Role: Develop NemRac and maintain it.
Research interests: Deep learning and its applications.
Contact: xxj@njau.edu.cn